Publications
Our Lab
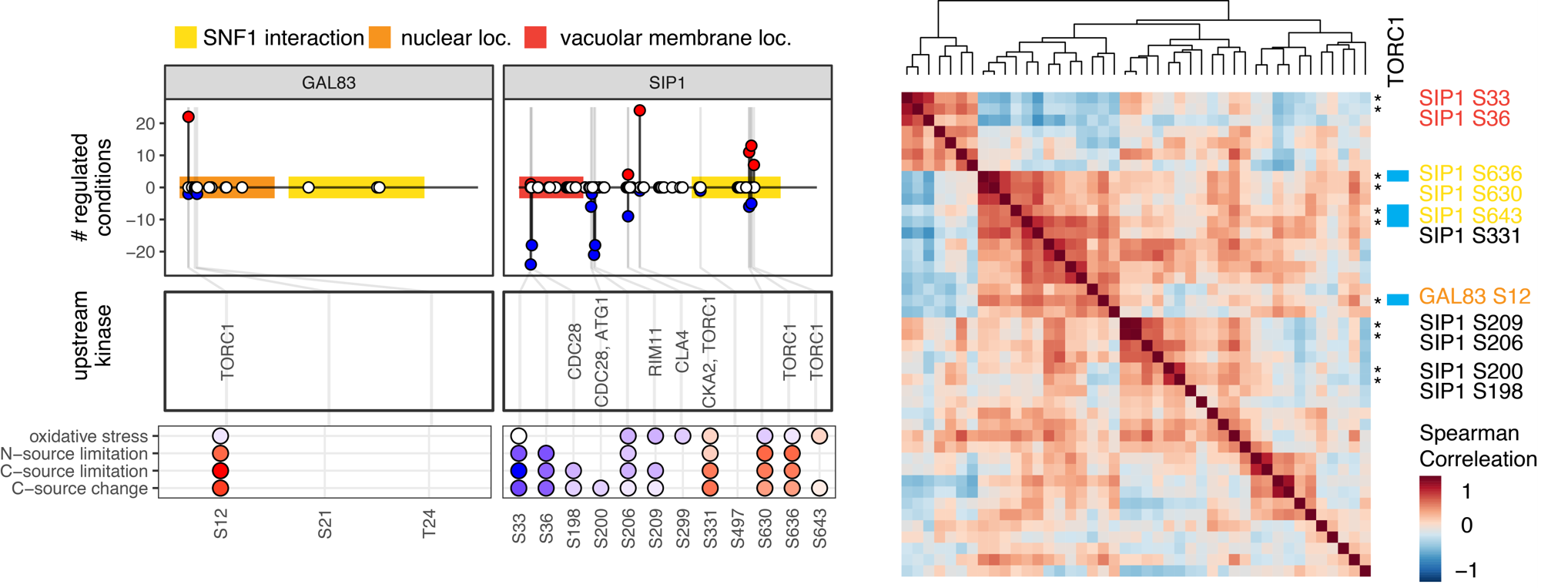
Mario Leutert, Anthony S. Barente, Noelle K. Fukuda, Ricard A. Rodriguez-Mias, Judit Villén. The regulatory landscape of the yeast phosphoproteome. Nat Struct Mol Biol 2023 30(11):1761-1773; doi: 10.1038/s41594-023-01115-3
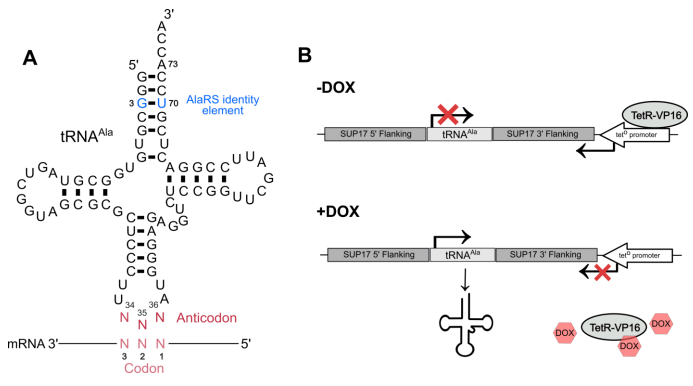
Ecaterina Cozma, Megha Rao, Madison Dusick, Julie Genereaux, Ricard A. Rodriguez-Mias, Judit Villén, Christopher J. Brandl, Matthew D. Berg. Anticodon sequence determines the impact of mistranslating tRNAAla variants. bioRxiv 2022.11.23.517754; doi: 10.1101/2022.11.23.517754
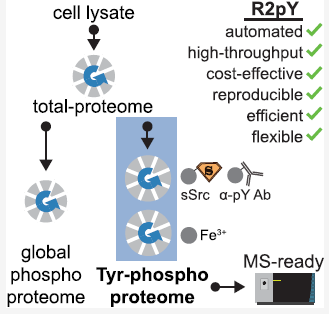
Alexis Chang, Mario Leutert, Ricard A. Rodriguez-Mias, Judit Villén. Automated Enrichment of Phosphotyrosine Peptides for High-Throughput Proteomics. Journal of Proteome Research 2023 22 (6), 1868-1880. doi: 10.1021/acs.jproteome.2c00850
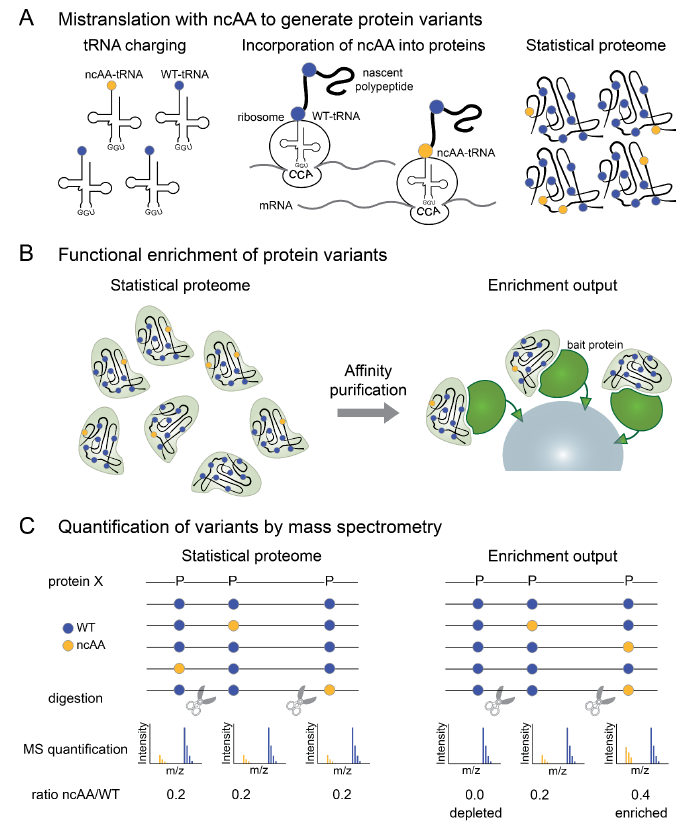
Ricard A. Rodriguez-Mias, Kyle N. Hess, Bianca Y. Ruiz, Ian R. Smith, Anthony S. Barente, Stephanie M. Zimmerman, Yang Y. Lu, William S. Noble, Stanley Fields, Judit Villén. Proteome-wide identification of amino acid substitutions deleterious for protein function. bioRxiv 2022.04.06.487405; doi: 10.1101/2022.04.06.487405
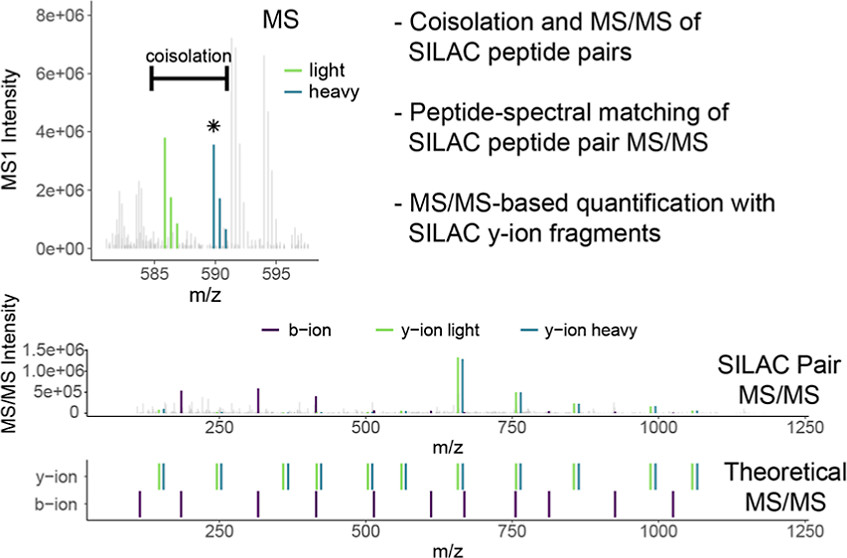
Smith IR, Eng JK, Barente AS, Hogrebe A, Llovet A, Rodriguez-Mias RA, Villén J. Coisolation of Peptide Pairs for Peptide Identification and MS/MS-Based Quantification. Anal Chem. 2022 Nov 8;94(44):15198-15206. doi: 10.1021/acs.analchem.2c01711.
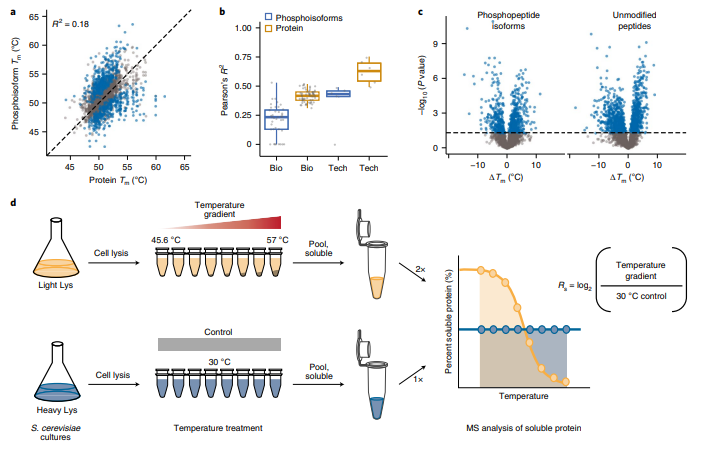
Smith IR, Hess KN, Bakhtina AA, Valente AS, Rodríguez-Mias RA, Villén J. Identification of phosphosites that alter protein thermal stability. Nat Methods. 2021 Jul;18(7):760-762. doi: 10.1038/s41592-021-01178-4.
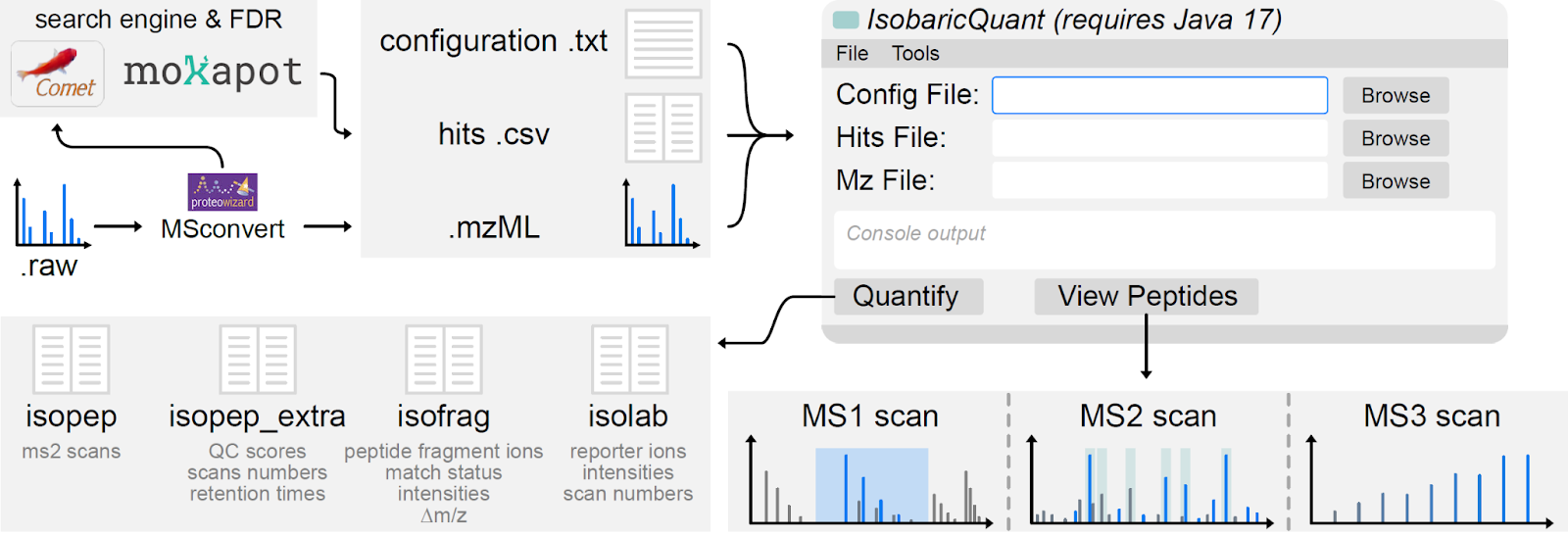
Hogrebe A, Hess KN, Llovet A, Ramos YJ, Barente AS, Hernandez-Portugues D, Smith IR, Rodríguez-Mias RA, Villén J. IsobaricQuant enables cross-platform quantification, visualization, and filtering of isobarically-labeled peptides. Proteomics. 2022 Oct;22(19-20):e2100253. doi: 10.1002/pmic.202100253.
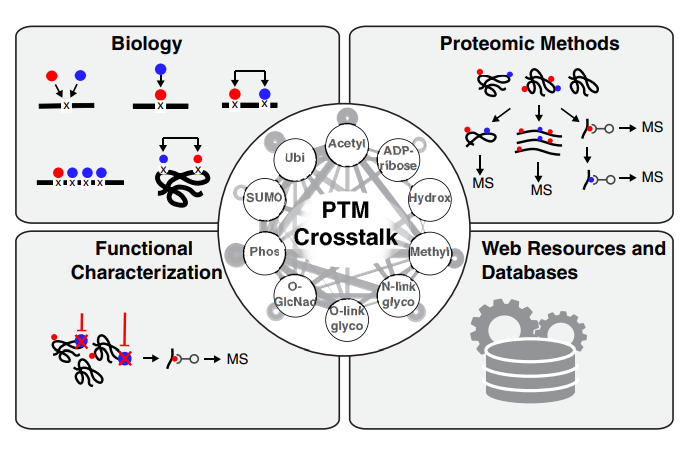
Leutert M, Entwisle SW, Villén J. Decoding Post-Translational Modification Crosstalk With Proteomics. Mol Cell Proteomics. 2021;20:100129. doi: 10.1016/j.mcpro.2021.100129.
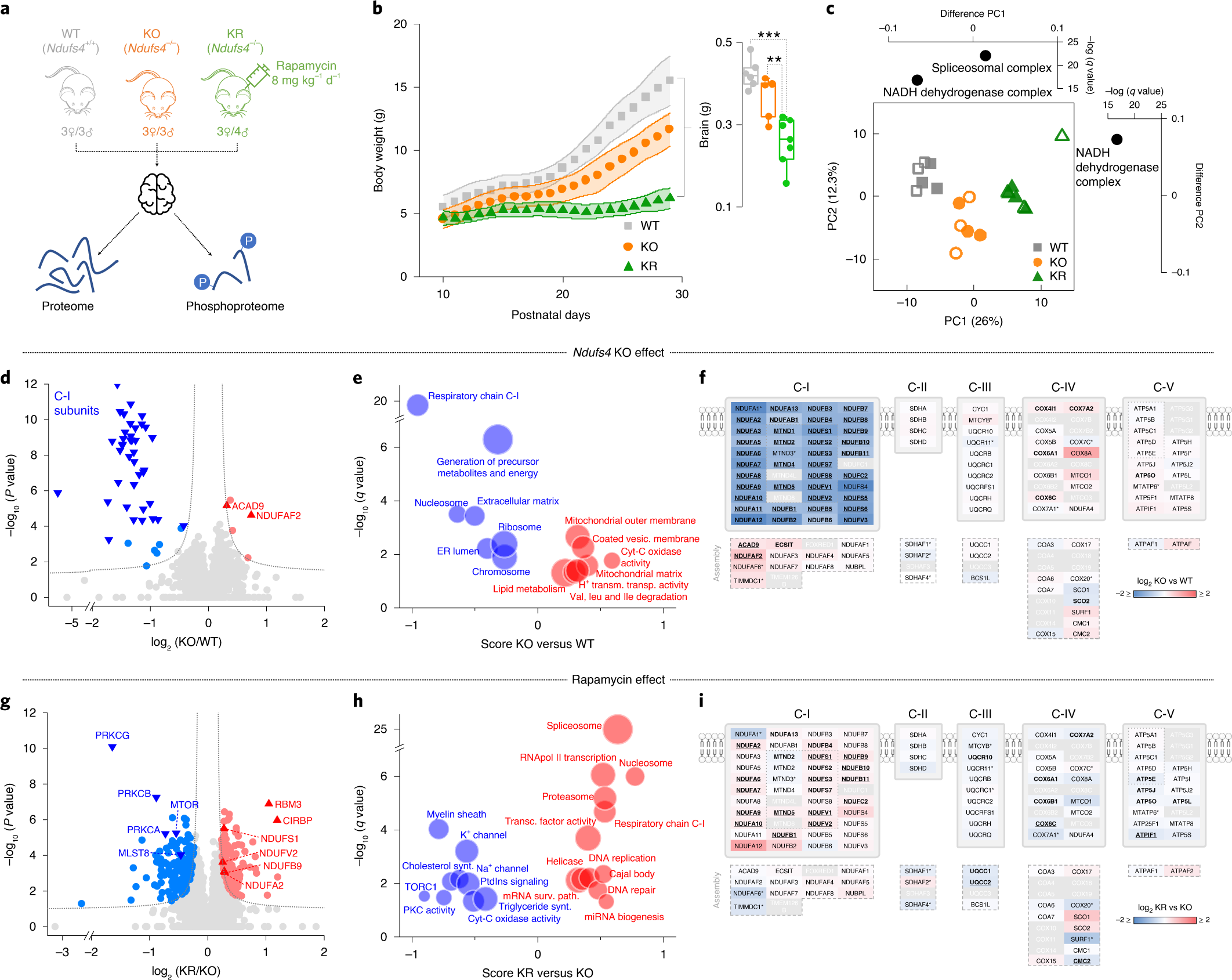
Martin-Perez M, Grillo AS, Ito TK, Valente AS, Han J, Entwisle SW, Huang HZ, Kim D, Yajima M, Kaeberlein M, Villén J. PKC downregulation upon rapamycin treatment attenuates mitochondrial disease. Nat Metab. 2020 Dec;2(12):1472-1481. doi: 10.1038/s42255-020-00319-x.
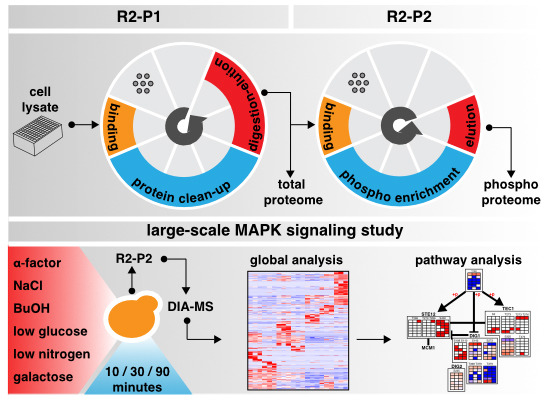
Leutert M, Rodríguez-Mias RA, Fukuda NK, Villén J. R2-P2 rapid-robotic phosphoproteomics enables multidimensional cell signaling studies. Mol Syst Biol. 2019 Dec;15(12):e9021. doi: 10.15252/msb.20199021.
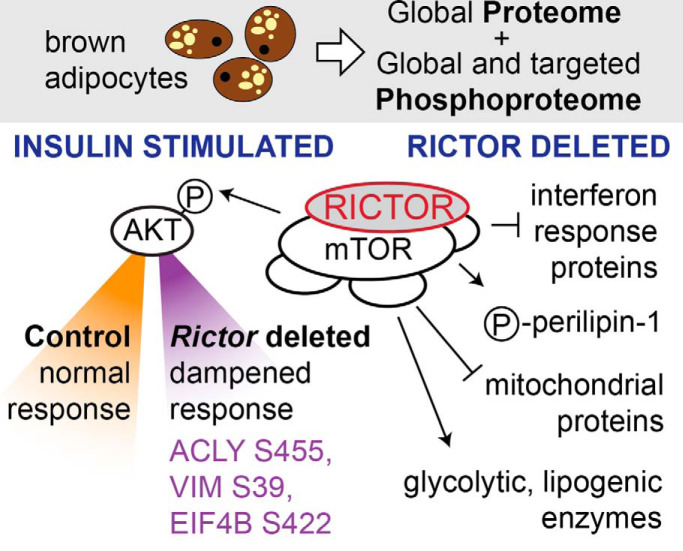
Entwisle SW, Martinez Calejman C, Valente AS, Lawrence RT, Hung CM, Guertin DA, Villén J. Proteome and Phosphoproteome Analysis of Brown Adipocytes Reveals That RICTOR Loss Dampens Global Insulin/AKT Signaling. Mol Cell Proteomics. 2020 Jul;19(7):1104-1119. doi: 10.1074/mcp.RA120.001946.
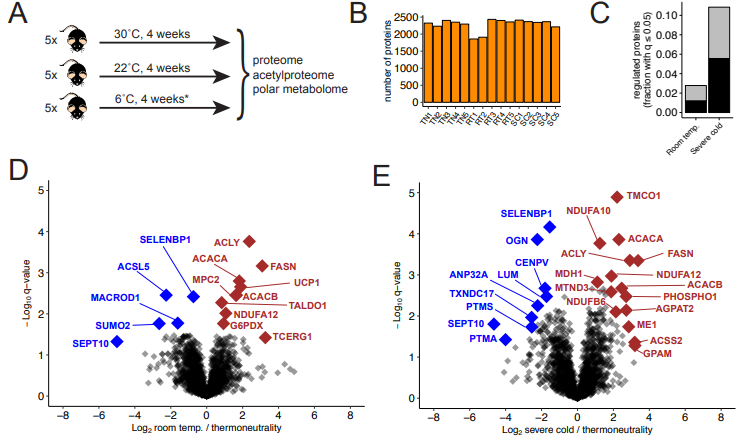
Entwisle SW, Sanchez-Gurmaches J, Lawrence RT, Pedersen DJ, Jung SM, Martin-Perez M, Guilherme A, Czech MP , Guertin DA, Villen J. Cold-Induced Thermogenesis Increases Acetylation on the Brown Fat Proteome and Metabolome. bioRxiv 445718; doi: 10.1101/445718
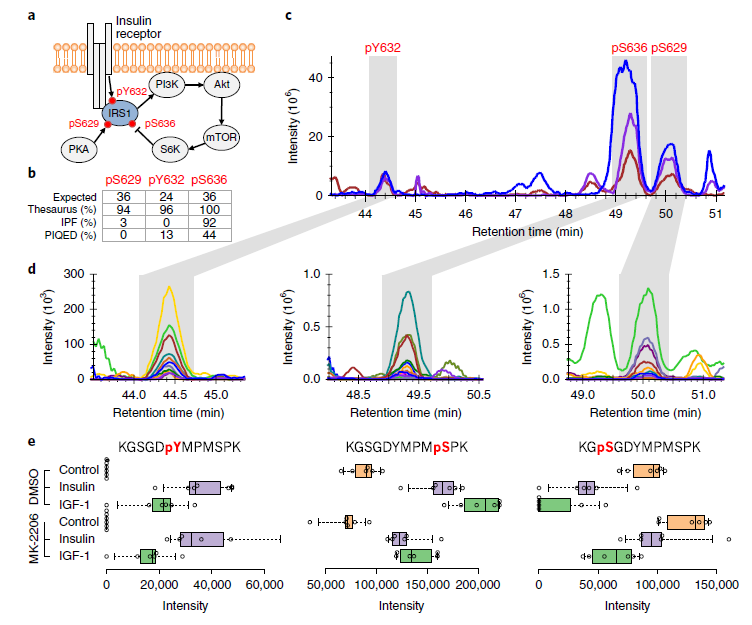
Searle BC, Lawrence RT, MacCoss MJ, Villén J. (2019) Thesaurus: quantifying phosphopeptide positional isomers. Nat Methods. 16(8):703-706. doi: 10.1038/s41592-019-0498-4.
Martin-Perez M and Villén J. (2017) Determinants and Regulation of Protein Turnover in Yeast. Cell Systems. Cell Systems 5, 283–294. doi: 10.1016/j.cels.2017.08.008
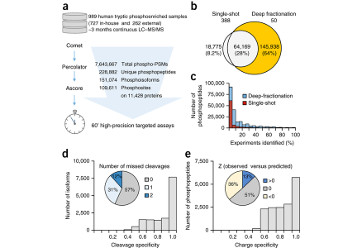
Lawrence RT, Searle BC, Llovet A and Villén J. Plug and play analysis of the human phosphoproteome by targeted high-resolution mass spectrometry. Nature Methods (2016) Mar 28. doi: 0.1038/nmeth.3811.
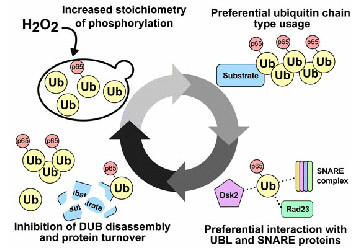
Swaney DL, Rodríguez-Mias RA, Villén J. (2015). Phosphorylation of ubiquitin at Ser65 affects its polymerization, targets, and proteome-wide turnover. EMBO Rep. 16,1131–1144.
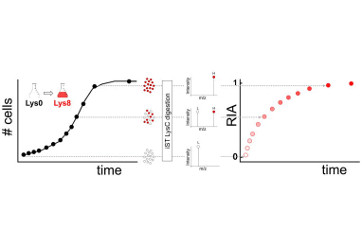
Martin-Perez, M., and Villen, J. (2015). Feasibility of protein turnover studies in prototroph S. cerevisiae strains. Anal. Chem. 87, 4008–4014.
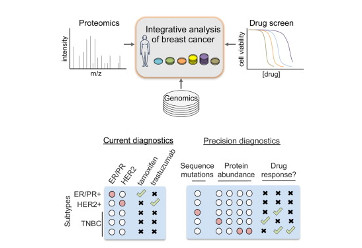
Lawrence RT, Perez EM, Hernández D, Miller CP, Haas KM, Irie HY, Lee SI, Blau CA, Villén J. (2015). The proteomic landscape of triple-negative breast cancer. Cell Rep. 11, 630–644.
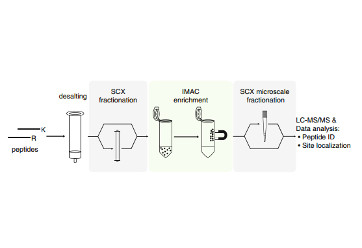
Edelman, W.C., Haas, K.M., Hsu, J.I., Lawrence, R.T., and Villen, J. (2014). A practical recipe to survey phosphoproteomes. Methods Mol. Biol. 1156, 389–405.
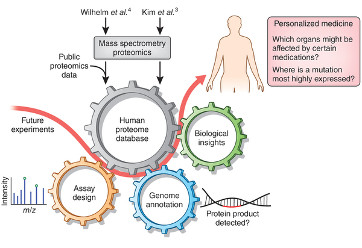
Lawrence, R.T., and Villen, J. (2014). Drafts of the human proteome. Nat. Biotechnol. 32, 752–753.
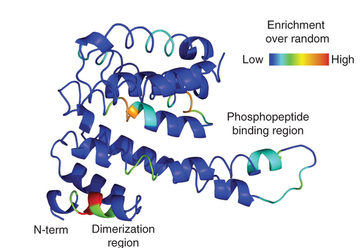
Swaney, D.L., Beltrao, P., Starita, L., Guo, A., Rush, J., Fields, S., Krogan, N.J., and Villen, J. (2013). Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nature Methods 10, 676–682. PDF Supplementary_info RAW data
Collaborations
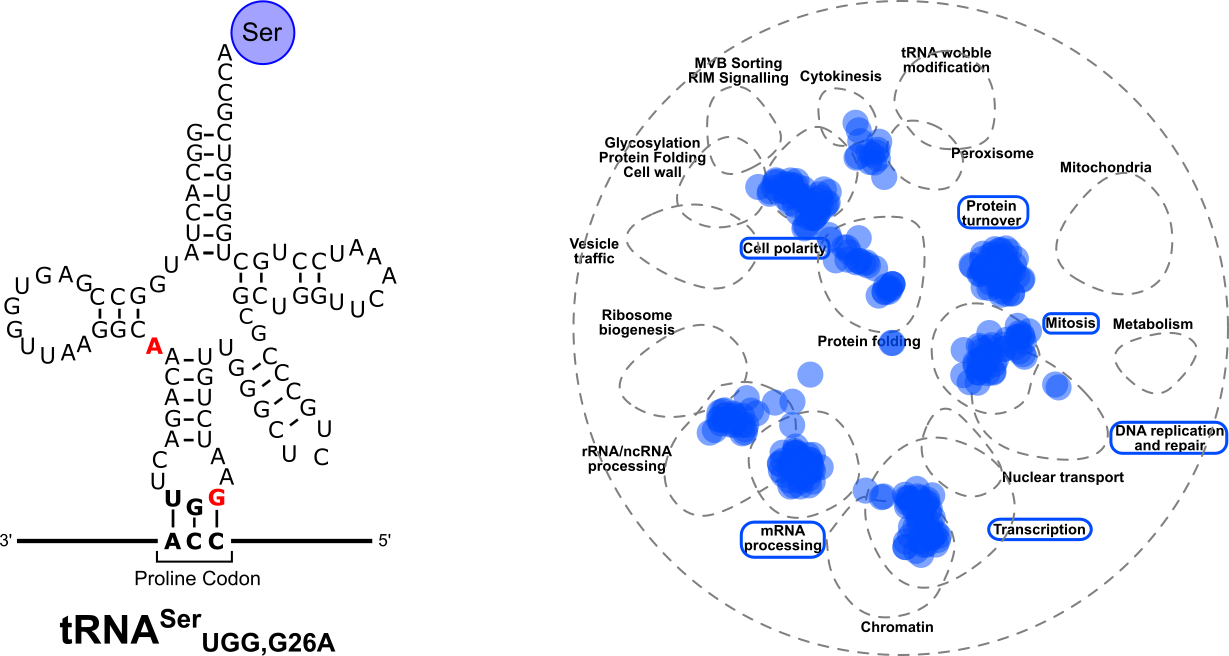
Berg MD, Zhu Y, Loll-Krippleber R, San Luis BJ, Genereaux J, Boone C, Villén J, Brown GW, Brandl CJ. Genetic background and mistranslation frequency determine the impact of mistranslating tRNASerUGG. G3 (Bethesda). 2022 Jul 6;12(7):jkac125. doi: 10.1093/g3journal/jkac125.

Isaacson JR, Berg MD, Charles B, Jagiello J, Villén J, Brandl CJ, Moehring AJ. A novel mistranslating tRNA model in Drosophila melanogaster has diverse, sexually dimorphic effects. G3 (Bethesda). 2022 May 6;12(5):jkac035. doi: 10.1093/g3journal/jkac035.
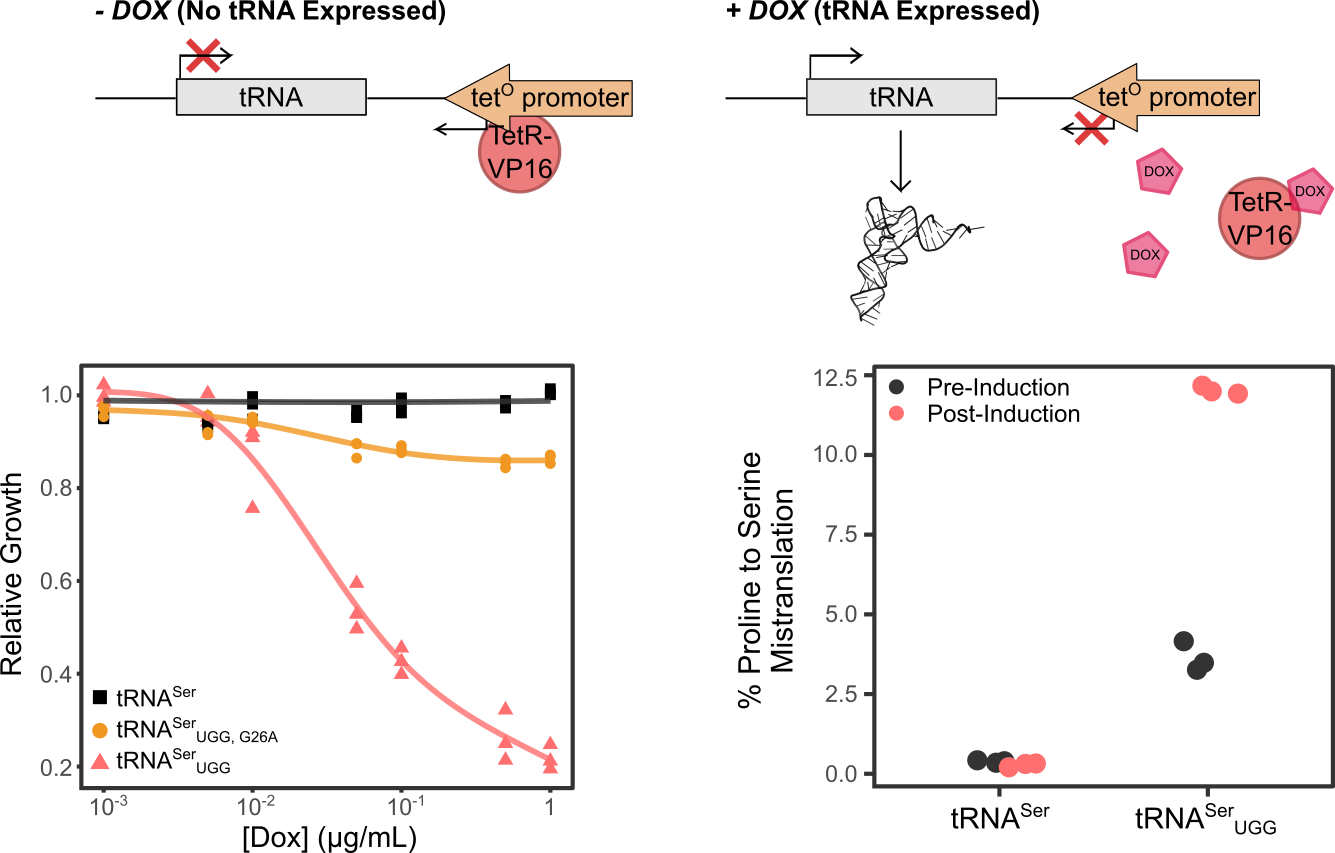
Berg MD, Isaacson JR, Cozma E, Genereaux J, Lajoie P, Villén J, Brandl CJ. Regulating Expression of Mistranslating tRNAs by Readthrough RNA Polymerase II Transcription. ACS Synth Biol. 2021 Nov 19;10(11):3177-3189. doi: 10.1021/acssynbio.1c00461.
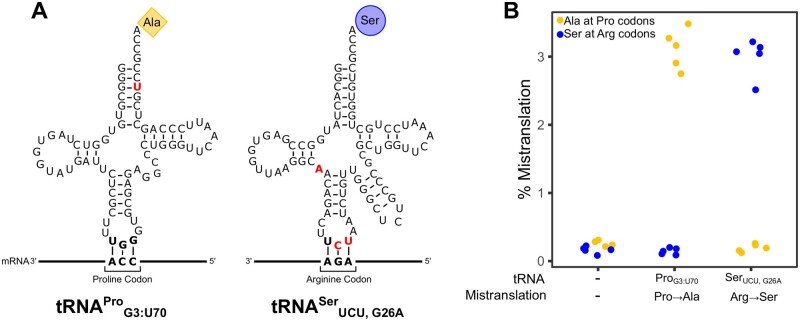
Berg MD, Zhu Y, Ruiz BY, Loll-Krippleber R, Isaacson J, San Luis BJ, Genereaux J, Boone C, Villén J, Brown GW, Brandl CJ. The amino acid substitution affects cellular response to mistranslation. G3 (Bethesda). 2021 Sep 27;11(10):jkab218. doi: 10.1093/g3journal/jkab218.
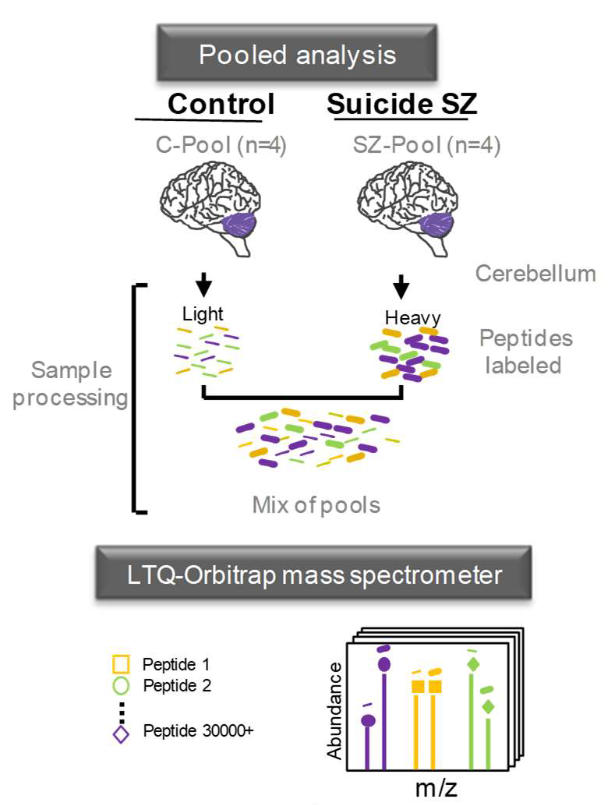
Vidal-Domènech F, Riquelme G, Pinacho R, Rodriguez-Mias R, Vera A, Monje A, Ferrer I, Callado LF, Meana JJ, Villén J, Ramos B. Calcium-binding proteins are altered in the cerebellum in schizophrenia. PLoS One. 2020 Jul 8;15(7):e0230400. doi: 10.1371/journal.pone.0230400.
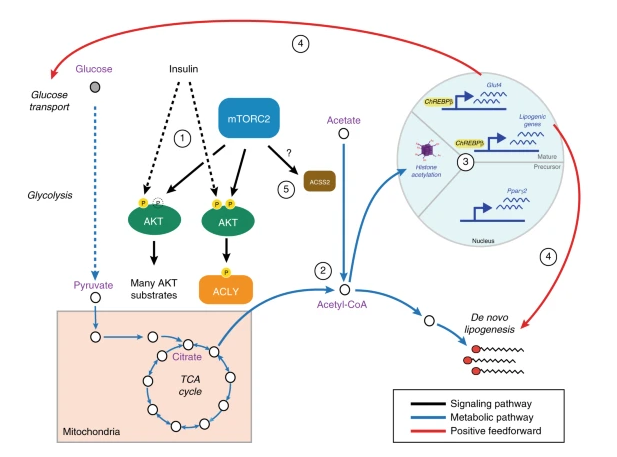
Martinez Calejman C, Trefely S, Entwisle SW, Luciano A, Jung SM, Hsiao W, Torres A, Hung CM, Li H, Snyder NW, Villén J, Wellen KE, Guertin DA. mTORC2-AKT signaling to ATP-citrate lyase drives brown adipogenesis and de novo lipogenesis. Nat Commun. 2020 Jan 29;11(1):575. doi: 10.1038/s41467-020-14430-w.
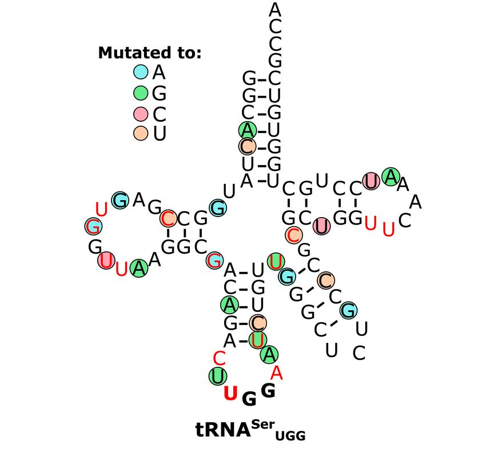
Berg MD, Zhu Y, Genereaux J, Ruiz BY, Rodriguez-Mias RA, Allan T, Bahcheli A, Villén J, Brandl CJ. (2019). Modulating mistranslation potential of tRNASer in Saccharomyces cerevisiae. GENETICS 213(3):849-863.
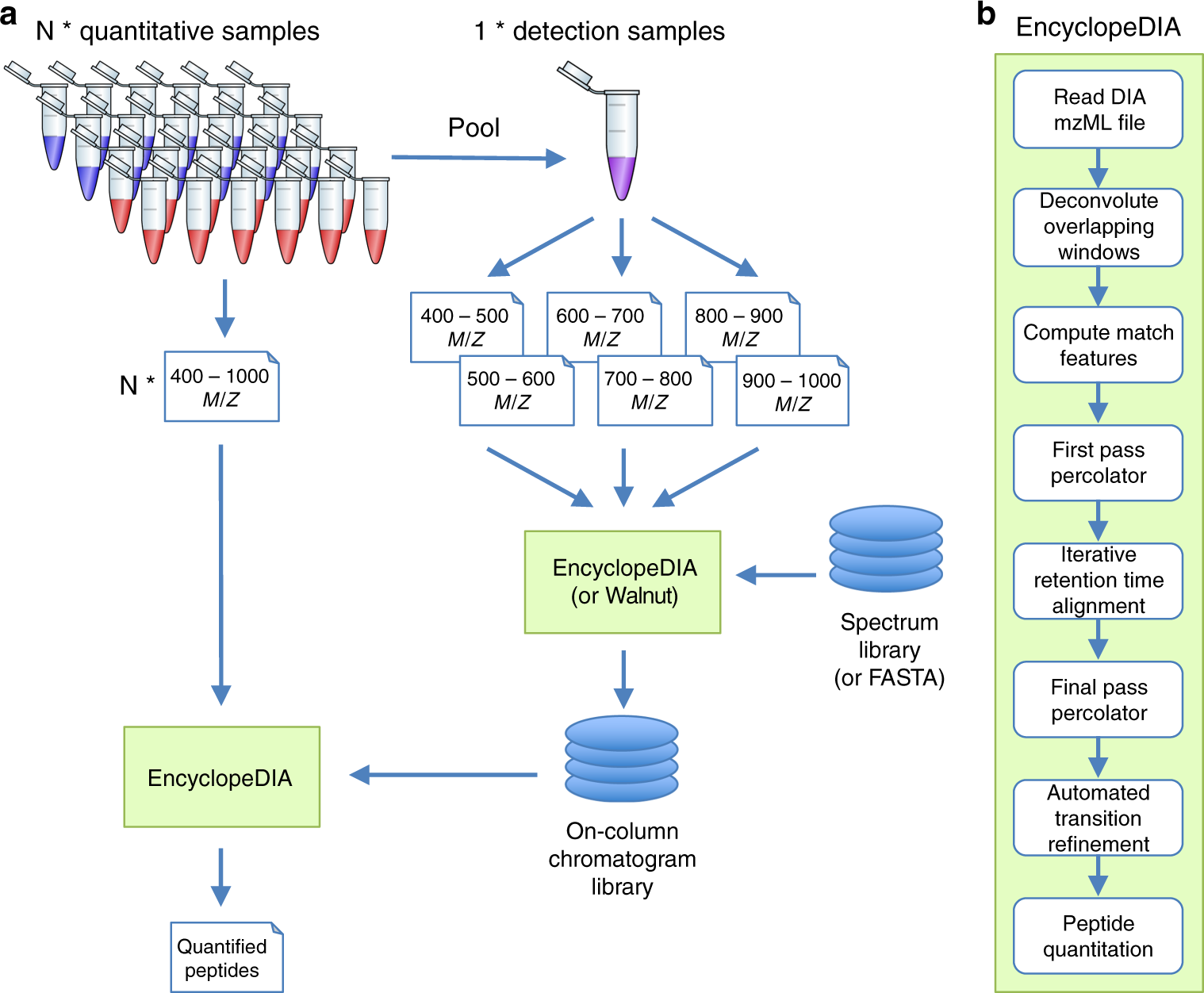
Searle BC, Pino LK, Egertson JD, Ting YS, Lawrence RT, MacLean BX, Villén J, MacCoss MJ. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat Commun. 2018 Dec 3;9(1):5128. doi: 10.1038/s41467-018-07454-w.
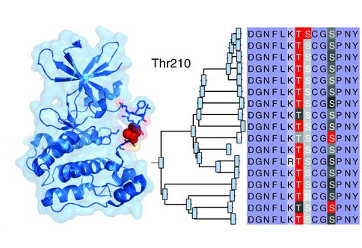
Studer RA, Rodriguez-Mias RA, Haas KM, Hsu JI, Viéitez C, Solé C, Swaney DL, Stanford LB, Liachko I, Böttcher R, Dunham MJ, de Nadal E, Posas F, Beltrao P, Villén J. Evolution of protein phosphorylation across 18 fungal species. Science 2016 Oct 14;354(6309):229-232.
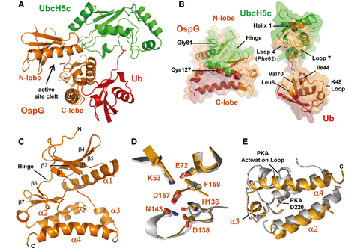
Pruneda, J.N., Smith, F.D., Daurie, A., Swaney, D.L., Villen, J., Scott, J.D., Stadnyk, A.W., Le Trong, I., Stenkamp, R.E., Klevit, R.E., et al. (2014). E2~Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis. EMBO J. 33, 437–449.
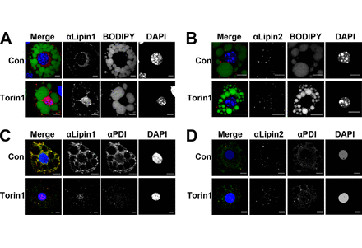
Eaton, J.M., Takkellapati, S., Lawrence, R.T., McQueeney, K.E., Boroda, S., Mullins, G.R., Sherwood, S.G., Finck, B.N., Villen, J., and Harris, T.E. (2014). Lipin 2 binds phosphatidic acid by the electrostatic-hydrogen bond switch mechanism independent of phosphorylation. J. Biol. Chem. 289, 18055-66
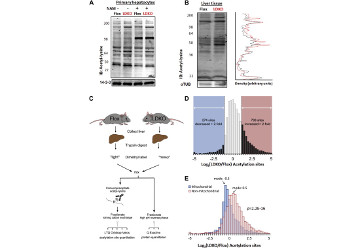
Chow, J.D.Y., Lawrence, R.T.*, Healy, M.E., Dominy, J.E., Liao, J.A., Breen, D.S., Byrne, F.L., Kenwood, B.M., Lackner, C., Okutsu, S., Mas, V.R., Caldwell, S.H., Tomsig, J.L., Cooney, G.J., Puigserver, P.B., Turner, N., James, D.E., Villén, J.#, and Hoehn, K.L.# (2014). Genetic inhibition of hepatic acetyl-CoA carboxylase activity increases liver fat and alters global protein acetylation. Mol. Metab. 3, 419–431. (*equal contribution; #co-corresponding)
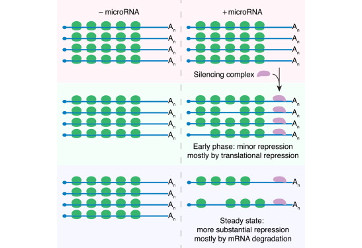
Eichhorn, S.W., Guo, H., McGeary, S.E., Rodríguez-Mias, R.A., Shin, C., Baek, D., Hsu, S.-H., Ghoshal, K., Villen, J., and Bartel, D.P. (2014). mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 56, 104–115.
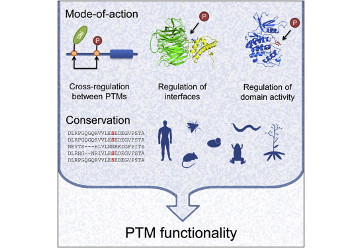
Beltrao, P., Albanèse, V., Kenner, L.R., Swaney, D.L., Burlingame, A., Villen, J., Lim, W.A., Fraser, J.S., Frydman, J., and Krogan, N.J. (2012). Systematic functional prioritization of protein posttranslational modifications. Cell 150, 413–425.
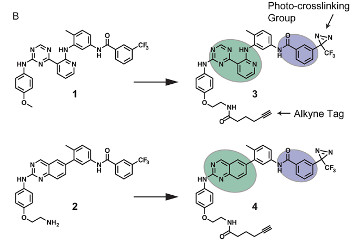
Ranjitkar, P., Perera, B.G.K., Swaney, D.L., Hari, S.B., Larson, E.T., Krishnamurty, R., Merritt, E.A., Villen, J., and Maly, D.J. (2012). Affinity-based probes based on type II kinase inhibitors. J. Am. Chem. Soc. 134, 19017–19025.
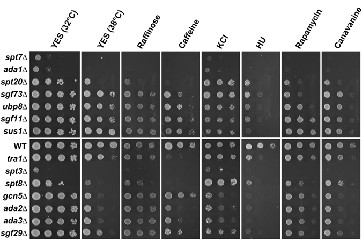
Helmlinger, D., Marguerat, S., Villen, J., Swaney, D.L., Gygi, S.P., Bähler, J., and Winston, F. (2011). Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. EMBO J. 30, 2843–2852